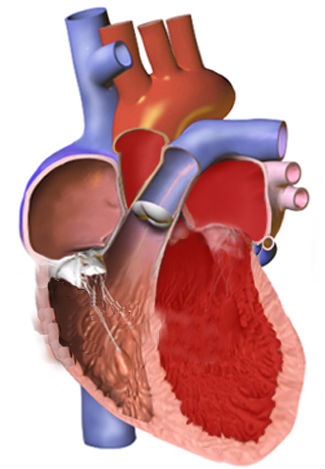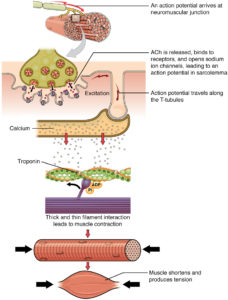
Previous studies reported that the formation and maintenance of the junctional membrane complexes, and thus the transverse tubule network (Figure 2), depends on JPH2. Furthermore, JPH2 interacts with RyR2 to hold the channel in a closed state (2), Quick and colleagues (1) used mass spectrometry-based proteomic analysis of the proteins that coimmunoprecipitated with either JPH2 or RyR2 from adult mouse hearts under relatively stringent immunoprecipitation conditions. This analysis identified SPEG as the only common binding partner for both proteins.
Genetic variants in each of the three genes, JPH2, RYR2, and SPEG, are associated with congenital heart disease (OMIM: JPH2, SPEG, RYR2). JPH2 and SPEG are associated with cardiomyopathy and RYR2 is associated with arrhythmias. Additionally, altered or reduced function of JPH2 and RyR2 is associated with heart failure and arrhythmias (2).
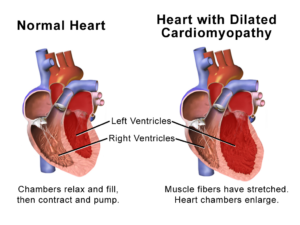
To discover how SPEG affects heart function, the authors developed a genetically engineered mouse model in which they could induce loss of SPEG expression specifically in heart cells. Inducing this reduction in SPEG abundance in adult mice resulted in heart failure that was associated with dilation of the ventricular heart tissue (dilated cardiomyopathy), fibrosis of the atrial heart tissue, disruption of the junctional membrane complexes leading to loss of the transverse tubule network in the cardiac muscle cells, and reduced calcium release in cardiomyocytes that was consistent with impaired excitation-contraction coupling.
Disruption of the transverse tubule network occurred before most changes in calcium signaling and were associated with a decrease in the amount of phosphorylated JPH2 but not the overall amount of JPH2. Thus, reducing SPEG abundance resulted in less phosphorylation of JPH2 and destabilization of junctional membrane complexes. This likely produced the impaired excitation-contraction coupling, which resulted in the compensatory changes of ventricular dilation and atrial fibrosis that occurred and then contributed to the lethality of the heart failure in the SPEG-deficient mice.
This study shows that targeted proteomic analyses can provide insight into human disease by revealing previously unknown interactions between proteins. On the basis of the proteomic data, the subsequent research identified a link between SPEG abundance and function, excitation-contraction coupling, and subcellular organization of cardiac muscle cell transverse tubule network. The proteomic data will lead cardiovascular research into new areas, especially in understanding the function of SPEG in heart failure.
Highlighted Articles
- A. P. Quick, Q. Wang, L. E. Phiippen, G. Barreto-Torres, D. Y. Chiang, D. Beavers, G. Wang, M. Khalid, J,. O,. Reynolds, H. M. Campbell, J. Showell, M. D. McCauley, A. Scholten, X. H. T. Wehrens, SPEG (striated muscle preferentially expressed protein kinase) is essential for cardiac function by regulating junctional membrane complex activity. Circ. Res. 120, 110-119 (2017). PubMed
- D. L. Beavers, A. P. Landstrom, D. Y. Chiang, X. H. T. Wehrens, Emerging roles of junctophilin-2 in the heart and implications for cardiac diseases. Cardiovasc. Res.103, 198–205 (2014). PubMed
Cite as: N. R. Gough, Proteomic Analysis Leads to Insights into Heart Failure. BioSerendipity (20 June 2017). https://www.bioserendipity.com/2017/06/20/proteomic-analysis-leads-to-insights-into-heart-failure/