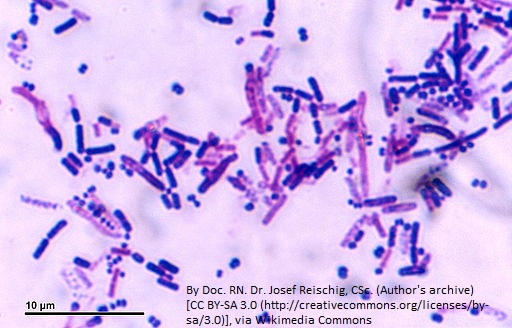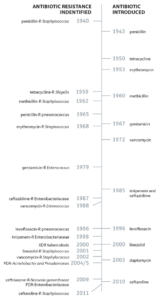
Antibiotic-resistant bacteria are becoming a major threat to human health (Figure 1). After a recent scare with a relative who contracted pneumonia while in the hospital, I began to think about antibiotic resistance and how bacteria become resistant even to those antibiotics that are rarely used, such as the carbapenem antibiotics.
Bacteria can be infected themselves with viruses, which are called bacteriophage. Bacteria contain genetic material in the form of chromosomes, and they often also contain extrachromosomal genetic material, called plasmids. Bacteria can exchange genetic material within the same species and between different species. These three properties, along with genetic mutations, give bacteria the ability to rapidly become resistant to antibiotics.
Humans are constantly being exposed to bacteria. Most are harmless or even beneficial, but some cause disease. The disease-causing ones are called “pathogenic.” The immune system effectively eliminates most of the pathogenic bacteria so that symptomatic infection does not happen. Infection arises when the pathogen divides and infects cells faster than the immune system can control the pathogen. This is when antibiotics are needed.
Antibiotics come in two main types: Those that kill bacteria (bactericidal) and those that stop the bacteria from dividing (bacteriostatic). A bacteriostatic antibiotic gives the immune system a chance to catch up and kill the bacteria and infected cells. Antibiotics do not discriminate between the good and the pathogenic bacteria. For most situations, either type of antibiotic will cure an infection unless the bacteria have become resistant.
Mutations in individual genes can render a bacterial species or subspecies strain resistant to an antibiotic. Bacteria can also become resistant by acquiring genes that mediate resistance from other bacteria. This transfer of genetic material can occur through three processes collectively referred to as horizontal gene transfer:
- transformation, shedding of the extrachromosomal DNA, which is then taken up by nearby bacteria,
- conjugation, direct transfer of DNA through a connection made between two bacteria, or
- transduction, transfer of DNA mediated by a bacteriophage.
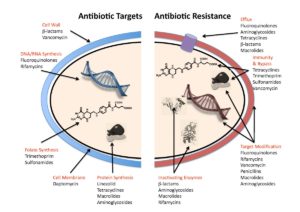
Bacteria can become resistant in multiple ways (Figure 2):
- by preventing the antibiotic from getting inside,
- by actively pumping the antibiotic out,
- by inactivating the antibiotic through a biochemical modification
- by degrading the antibiotic, or
- by modifying the molecular target of the antibiotic.
Because bacteria divide rapidly, mutations can occur and spread through bacteria more quickly than in a slower growing organism. The presence of antibiotic will naturally select those bacteria that gain mutations enabling them to continue to grow and divide in the presence of the antibiotic. For a mutation that affects the molecular target of the antibiotic, then an antibiotic with a different mechanism of action would still be effective if the immune system could not clear the residual infection caused by the mutant bacteria. This type of mutation would not typically cause multidrug resistance, that is resistance to multiple antibiotics.
Multidrug resistance could happen by the acquisition of genes or mutations that prevent antibiotics from reaching their molecular targets, for example genes or mutations that block antibiotic entry or enhance antibiotic efflux. Multidrug resistance could also occur through the acquisition of enzymes that modify or inactivate multiple types of antibiotics. For example, the presence of genes, either on the bacterial chromosome or on a plasmid, for various types of beta-lactamase enzymes results in resistance to specific classes of penicillin-type antibiotics. The presence of the gene SHV-1 results in resistance to multiple beta-lactam-based antibiotics: ampicillin, piperacillin, tigecycline, and carbapenem. SHV-1 is present in the chromosome of some bacteria and on extrachromosomal plasmids in others. Multidrug resistance can occur through multiple steps, such as a chromosomal mutation of the molecular target of one antibiotic, followed by increased production of a drug efflux pump that causes resistance to another set of antibiotics, followed by acquisition of a plasmid encoding a protein that degrades yet another type of antibiotic, and so on, until the bacteria is resistant to all of the available antibiotics.
The ability of bacteria to evolve resistance in distinct ways is highlighted by the study by DeLeo and colleagues. They compared samples isolated from 83 patients to two reference strains ST258 of Klebsiella pneumoniae, which cause carbapenem-resistant respiratory infections. Their analysis showed that the bacteria had variations in the areas of the bacterial chromosome that came from bacteriophages (called prophage), which are like “latent” forms of eukaryotic viral infections. These regions could mediate the transfer of genetic material to bacteria subsequently infected with the bacteriophage and indicate that the bacteria had been infected by a bacteriophage, which could have transmitted resistance or introduced a resistance-triggering change in the chromosome.
Another region of the bacterial chromosome appeared to be a hotspot for genetic alterations. The differences in the chromosomal mutational “hot spot” were associated with genes involved in making the capsule (the polysaccharide coating on the cell wall), which suggested that these bacteria could have different antigens and stimulate different immune responses.
The authors also found that the bacteria had different numbers of plasmids with different combinations of resistance genes. Phylogenetic analysis to determine how closely related the 85 bacteria were revealed that they separated into two groups with plasmids encoding two different enzymes responsible for carbapenem resistance and with differences in the chromosomal hot spot. This analysis showed that resistance evolved in at least two ways; there was not a single strain of Klebsiella pneumoniae that gave rise to the multidrug-resistant strains.
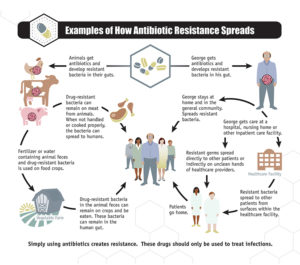
The prevalence of resistance-conveying bacteriophage in the food supply is demonstrated by Shousha and colleagues, who found that all but 1 of 50 chicken meat samples tested contained bacteriophage capable of infecting Escherichia coli. Random testing of the 243 phages showed that ~25% conferred resistance to one or more antibiotics when phage DNA was mixed with E. coli. Thus, antibiotic resistance may arise from the use of antibiotics in animals that are part of the food chain (Figure 3).
Thus, even antibiotics of last resort can have limited usefulness due to the acquisition of resistance through multiple mechanisms. The emergence of resistance can occur rapidly as plasmids or bacteriophage transfer genetic material among bacteria. Furthermore, resistance can spread into multiple bacteria species and strains and produce new combinations of resistance. As the antibiotics destroy the nonpathogenic “good” microbes, the resistant pathogenic bacteria grow with less competition, the immune system becomes overwhelmed, and a potentially lethal multidrug-resistant infection can result.
Related Literature
DeLeo, F.R., Chen, L., Porcella, S.F., Martens, C.A., Kobayashi, S.D., Porter, A.R., Chavda, K.D., Jacobs, M.R., Mathema, B., Olsen, R.J., Bonomo, R.A., Musser, J.M., Kreiswirth, B.N., Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc. Natl. Acad. Sci. U.S.A.111, 4988–4993 (2014). PubMed
Logan, L.K., Weinstein, R.A., The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J. Infect. Dis. 215, S28–S36 (2017). PubMed
Shousha, A., Awaiwanont, N., Sofka, D., Smulders, F.J.M., Paulsen, P., Szostak, M.P., Humphrey, T., Hilbert, F., Bacteriophages Isolated from Chicken Meat and the Horizontal Transfer of Antimicrobial Resistance Genes. Appl. Environ. Microbiol. 81, 4600–4606 (2015). PubMed
Ventola, C.L., 2015. The Antibiotic Resistance Crisis. Pharm. Ther. 40, 277–283 (2015). PubMed
Anderson, U. A Brief Review of Carbapenems. Pharmacy Times (2016). Full text
Related Online Resources
Carbapenems, MedScape. https://reference.medscape.com/drugs/carbapenems (accessed 24 September 2020)
Antibiotic/Antimicrobial Resistance, Centers for Disease Control and Prevention. https://www.cdc.gov/drugresistance/about.html (accessed 14 August 2017).
Microbiology Module, Antimicrobial Resistance Learning Site, Michigan State University. https://amrls.umn.edu/microbiology (accessed 14 August 2017)
Cite as: N. R. Gough, Resistance to Antibiotics of Last Resort. BioSerendipity (14 August 2017) https://www.bioserendipity.com/2017/08/14/resistance-to-antibiotics-of-last-resort/.