A Proteomic Approach to the Tumor Microenvironment
An outstanding issue in treating solid cancers is understanding the complexity of this pathological tissue. Solid tumors are comprised not only of the cancer cells, but they also contain immune cells, cells that form blood vessels and lymphatic vessels, fibroblasts, and the stem cells that form the fibroblasts and other nonimmune cells in the tumor (1). This complex pathological tissue is called the “tumor microenvironment” (Figure 1).
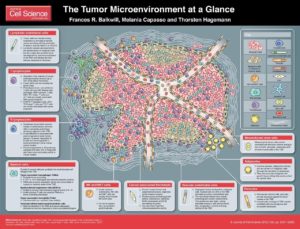
J Cell Sci 125, 5591-5596 (2012). doi: 10.1242/jcs.116392. Visit http://jcs.biologists.org/content/125/23/5591 for the full-size version.
The tumor microenvironment is necessary for a solid tumor to grow beyond a certain size, because as tumors grow larger they cannot obtain the necessary nutrients and exchange oxygen and waste products with the circulation. Furthermore, for most tumors, at some point the immune system detects the tumor cells or the injury and inflammation associated with the tumor, which leads to the recruitment of the immune cells.
This complex environment is partly why therapies that killed cancer cells so effectively in the lab failed to produce complete cures or durable remission when used in patients. Another reason is that the tumor cells themselves can be different from each other (heterogeneous) both in terms of genetic mutations and in terms of regulatory networks. Furthermore, cancer cells can evolve in the body in response to treatment. This change in the cancer cells can occur through additional genetic mutations that let the tumor resist a treatment. Cancer cells can also change without having new genetic mutations by rewiring their signaling, metabolic, and regulatory systems to survive in the presence of various therapeutic efforts to kill them. Cancer and treatments also trigger changes in the tumor microenvironment, To effectively eliminate cancer-associated death or produce a durable high quality of life with cancer, the complexity and evolving nature of not only the tumor cells but the tumor microenvironment need to be understood. A pair of papers describes the application of single-cell proteomics of dissociated tumor biopsy samples, through a technique called mass cytometry, to achieve better understanding of the complexity of the immune cell components of two types of tumors (2, 3) (Figure 2).
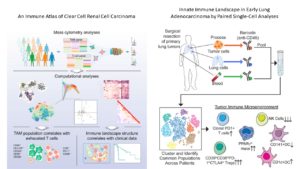
Chevrier and colleagues (2) examined the immune cell complement of clear cell renal carcinoma, a form of kidney cancer. They focused on two types of immune cells, one lymphoid (T cells) and one myeloid (macrophages). Their analysis identified 22 distinct types of T cells and 17 types of macrophages. With this information, the authors correlate progression-free survival with a specific immune cell profile, provide insights into the main protein markers of immunosuppression, and provide new insights into the complexity of the T cell and macrophage populations in a solid cancer.
Their analysis identified 22 distinct types of T cells and 17 types of macrophages.
Lavin and colleagues (3) tackled the difference between the immune cells in normal lung tissue, early stage lung adenocarcinoma, and peripheral blood from the same patient. In addition to mass cytometry, these authors analyzed the distribution of the immune cells in the tissue using a method called “multiplexed immunohistochemical consecutive staining on a single slide (MICSSS)” (4) and obtained transcriptomic data through a method called “massively parallel single-cell RNA-sequencing (MARS-seq)”. By integrating all three of these sets of data, the authors not only identified subsets of specific types of immune cells that were enriched or less abundant in tumor tissue compared to normal lung tissue, but they matched the position of these cells with specific regions of the tumor. Their analysis indicated that even early lung tumors had a profile of immunosuppression, indicating that as immune cells respond to tumors their profiles and behaviors change.
Even stage I lung tumors have a profile of immunosuppression.
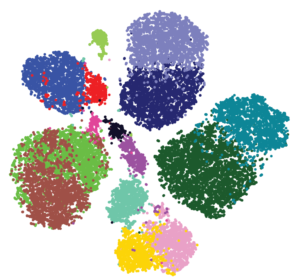
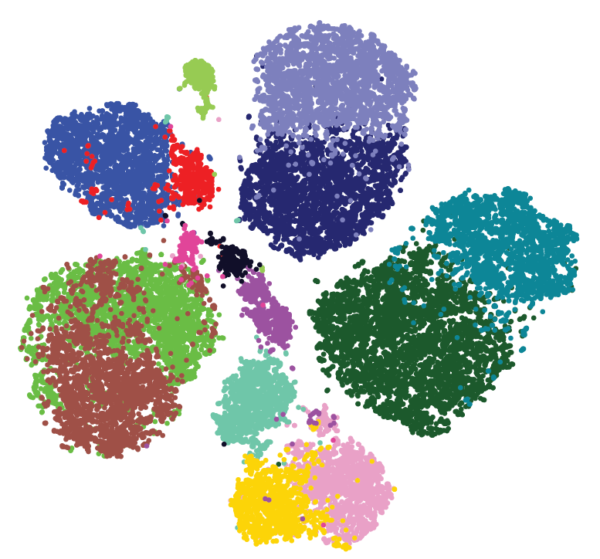
Both groups generated high-dimensional data and used a mathematical approach called Phenograph clustering to group the cells into subsets based on the similarities in their proteomic profiles (Figure 3). Together, these studies provide key insights that will lead to better understanding of which patients are most likely to benefit from immunomodulatory therapy. Understanding the changes that occur in the immune system at the early stages of cancer may also lead to the development of new types of anti-cancer immunotherapy or new approaches to keeping the immune system effective at eliminating cancer cells and preventing cancer from progressing.
Deep Dive into the Immune Cell Profiling Data of Kidney Cancer
Chevrier and colleagues (2) analyzed cells isolated from clear cell renal cell carcinoma (a form of kidney cancer) samples from 73 patients with cancers of different grades and from 5 samples of normal kidney (Figure 4). They took a targeted approach that used antibodies recognizing proteins that were present on various types of immune cells. A few of the antibodies recognized proteins on natural killer (NK) cells and mature or immature B cells. Most of the antibodies recognized various proteins on T cells or tumor-associated macrophages. These cells were the focus of the study, which aimed to determine how many different subtypes of these cells were present in the tumors and if an immune profile based on these subpopulations would be useful to predict disease progression.
The results indicated that most of the cells were lymphoid-type cells: 51% of the immune cells were T cells, 4% were B cells, and 9% were NK cells. Myeloid cells comprised 31% of the immune cell population. Myeloid cells produce macrophages and other types of non-lymphoid immune cells. T cells are broadly divided into classes by the presence or absence of 2 proteins, CD4 and CD8. T cells with both proteins (“double positive”) are immature and, once stimulated, become either positive for CD4 or CD8 (“single positive”). The presence of other proteins indicates whether these single-positive cells are active and dividing, are memory cells that have been activated previously but are no longer dividing, are regulatory cells that limit the activity of other T cells, or have become exhausted and will not respond to stimulation. The application of Phenograph clustering divided the T cell population into 22 different types (T-0 through T-21) based on the relative abundance of specific proteins.
Similar analysis of the tumor-associated macrophages identified 17 types (M-0 through M-16) of these cells. The function and behavior of specific types of macrophages is still an area of investigation. However, M-5, M-11, M-12, and M-13 represented populations of macrophages that were only present in tumor samples and not in the normal samples.
Tumor samples with a high number of M-5 type macrophages also tended to have a high number of T-0 type exhausted CD8-positive T cells. Some of the tumor samples with high numbers of M-5 and T-0 cells also had many T-6 type regulatory CD4-positive T cells. This profile of abundant M-5, T-0, and T-6 cells would be consistent with an immunosuppressive profile, suggesting that the immune system may have initially responded robustly to these tumors and consequently became exhausted. The immune response to these tumors would be impaired.
To determine if the profiles of CD4 T cells, CD8 T cells, or tumor-associated macrophages showed distinctive patterns across the patients, the authors used mathematical analysis to cluster the patients with the profiles associated with each of the populations of the three types immune cells. This analysis showed that CD4-positive profiles were similar across all the patients’ samples, the different types of tumor-associated macrophages were variable across the samples, and the CD8-positive populations showed a distinguishing pattern. Tumors with a lot of the two CD8-positive subpopulations that were high in an immune checkpoint receptor PD-1 (populations T-0 and T-1) did not have many of the CD8-positive cells lacking PD-1 (populations T-4, T-5, and T-11).
The presence of cells with abundant PD-1 would be indicative of exhausted T cells that could not respond to the tumor. Finding that T cells appeared to exist in the tumors as either exhausted or not may be clinically useful. Detecting PD-1-positive T cells in tumor biopsies may indicate that these patients would respond to immune checkpoint inhibitor therapy targeting PD-L1/PD-1. Additional evidence from the immune cell analysis that supported this therapeutic approach comes from the analysis of the abundance of markers of T cell exhaustion and immunosuppression. PD-1 was the most consistently present of the immunosuppressive proteins on T cells. The others, CTLA-4, TIM-3, and 4-1BB, tended to be more variable in abundance. Thus, testing for PD-1 and then targeting the PD-L1/PD-1 pathway is likely to be the most useful immune checkpoint inhibitor option in those patients with tumors enriched in PD-1-positive immune cells.
Another mathematical analysis that found that patients with tumors having few M-5 macrophages and high numbers of M-11 or M-13 macrophages or both M-11 and M-13 tended to die sooner. Thus, a M-11 or M-13 high and M-5 low signature correlated with poor prognosis in this group of patients. The correlation with a low M-5 profile may seem counterintuitive because M-5 cells are associated with exhausted T cells and regulatory T cells, which would be expected to impair the immune response to the cancer cells. However, immunosuppression through T cell exhaustion and regulatory T cells is not the only mechanism by which tumor cells can evade the immune system. In the patients analyzed, the more significant attribute than the presence of cells associated with immunosuppression was the presence of the M-11 and M-13 class of macrophages. This finding should spark investigation into the functions of these M-11 and M-13 macrophages and suggests that additional immunomodulatory approaches may be beneficial. This finding also raises the question of the importance of M-11 and M-13 macrophages in other types of cancer or in other groups of patients with kidney cancer.
This finding should spark investigation into the functions of these M-11 and M-13 macrophages and their importance in other types of cancer
The results also suggested that in addition to PD-1, the protein CD38, which is an enzyme, may be a consistent marker of T cell exhaustion and a marker of immunosuppressive macrophages, and provides new insights into the different states of macrophages, at least some which appeared related to the changes macrophages undergo as they leave the circulation and enter tissues and others of which relate to immunosuppressive status. Finally, this study supports the emerging concept that the immune system is a continuum of cells in different states and thus far more complex than previously appreciated.
Deep Dive into the Immune Cell Profiling Data of Lung Cancer
In the second paper, Lavin and colleagues analyzed matched samples of normal lung (Figure 5), peripheral blood, and lung adenocarcinoma from 18 patients. This analysis showed that T and B cells (lymphocytes) were more abundant in the tumor than in normal lung and that NK cells were less abundant in the tumor than in normal tissue. Macrophages were present in similar amounts in both normal and tumor tissue.
By applying the Phenograph clustering method, the authors determined the properties of the different types of immune cells specifically associated with the tumor. The regulatory T cells were not only more abundant in the tumor than the normal tissue but those in the tumor had a different molecular profile from those in the normal tissue. Not only were the cytotoxic CD8-positive T cells and NK cells less abundant in the tumor, but the ones that were present were less toxic than those in the normal tissue or the blood. CD4-positive T cells and CD8-positive T cells that were PD-1-positive were only detected in the tumor. The properties of these immune cells in the early stage tumors were consistent with an immunosuppressed phenotype.
The authors also investigated the myeloid-derived immune cells that were present in tumors using a combination of the mass cytometry analysis and transcriptomic analysis. By comparing the profiles of cells in the tumors with those in normal blood, the authors identified a profile specific to the tumor-associated macrophages and showed that a specific subtype of dendritic cell was excluded from tumors. The immune profile of the early lung cancer patients was likely to produce an immunosuppressive environment. This profile consisted of a high amount of a subset of macrophages, a subset of dendritic cells, a subset of regulatory T cells, and exhausted T cells and a low abundance of NK cells and cytotoxic cells. Using the transcriptomic profiling data, the authors analyzed transcriptomic information from 515 lung adenocarcinoma patients and showed that patients with a high ratio of macrophages with the tumor-associated transcriptomic profile to macrophages with a normal lung transcriptomic profile had poorer survival. Comparison of stage I patients with those with stage II or stage III lung adenocarcinoma revealed that the cellular immune profile was similar in both the early and later stages with one exception; later stages had more neutrophils. Thus, most changes in the immune system appeared to occur early during the development of this type of lung cancer.
Two other aspects of this study provide information that could be clinically relevant. The MICSSS analysis revealed that tumors with a lymph structure called tertiary lymphoid structures had more T cells, which were close to the antigen-presenting dendritic cells, and fewer macrophages. This organization of immune cells suggests that these structures help T cells infiltrate and become activated within the tumor. Tumors lacking these structures tended to have an accumulation of immune cells, including PD-L1-positive macrophages, in the tissue surrounding the tumor rather than infiltrating the tumor.
Tertiary lymphoid structures may help T cells infiltrate and become activated within the tumor.
Together, these studies illustrate the amazing complexity of the immune system, which is apparent when many different proteins are analyzed simultaneously through proteomic techniques. Furthermore, the lung cancer data suggests that even early stage tumors are associated with a suppressed immune response. Both studies indicate an important role for tumor-associated macrophages in tumor progression, suggesting that targeting these cells may be useful therapeutically. The kidney cancer results present an exciting opportunity to investigate mechanisms by which macrophages can contribute to clinical outcome in ways other than by altering T cell activity.
Highlighted Articles
- F. R. Balkwill, M. Capasso, T. Hagemann. The tumor microenvironment at a glance. J. Cell Sci. strong>125, 5591 (2013). PubMed
- S. Chevrier, J. H. Levine, V. R. T. Zanotelli, K. Silina, D. Schulz, M. Bacac, C. H. Ries, L. Ailles, M. A. S. Jewett. H. Moch, M. van den Broek, C. Beisel, M. B .Stadler, C. Gedye, B. Reis, D. Pe’er. B. Bodenmiller, An immune atlas of clear cell renal cell carcinoma. Cell 169, 736–749 (2017). PubMed
- Y. Lavin, S. Kobayashi, A. Leader, E. D. Amir, N. Elefant, C. Bigenwald, R. Renmark, R. Sweeney, C. D. Becker, J. H. Levine, K. Meinhof, A. Chow, S. Kim-Shulze, A. Wolf, C. Medaglia, H. Li, J. A. Rytlewski, R. O,. Emerson, A. Solovyov, B. D. Greenbaum, C. Sanders, M. Vignali, M. B. Beasley, R. Flores, S. Gnjatic, D. Pe’er, A. Rahman, I. Amit, M. Merad. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell 169, 750–765 (2017). PubMed
- R. Remark, T. Merghoub, N. Grabe, G. Litjens, D. Damotte, J. D. Wolchok, M. Merad,
S. Gnjatic, In-depth tissue profiling using multiplexed immunohistochemical consecutive
staining on single slide. Sci. Immunol. 1, aaf6925 (2016). Journal
Featured Methods
PhenoGraph (accessed 27 June 2017) https://www.c2b2.columbia.edu/danapeerlab/html/phenograph.html
Exploring “Impossible” Science:CyTOF and mass-tag barcoding let Eli Zunder ask more questions and find new answers (accessed 30 May 2020) https://www.fluidigm.com/articles/exploring-impossible-science
MARS-Seq:
- D. A. Jaitin, et al., Massively parallel single-cell RNA-Seq for marker-free decomposition of tissues into cell types. Science 343, 776-779 (2014) DOI: 10.1126/science.343.6172.707-m
- H. Keren-Shaul, et al, MARS-seq2.0: an experimental and analytical pipeline for indexed sorting combined with single-cell RNA sequencing. Nature Protocols 14, 1841-1862 (2019) DOI: 10.1038/s41596-019-0164-4
MICSSS (accessed 27 June 2017) http://www.mountsinai.org/about-us/newsroom/press-releases/mount-sinai-researchers-develop-simple-method-to-characterize-immune-cells-in-tumors-
Cite as: N. R. Gough, Single-Cell Proteomics Shines Light on the Complexity of Immune Cells in Solid Tumors. BioSerendipity (28 June 2017) https://www.bioserendipity.com/single-cell-proteomics-shines-light-on-the-complexity-of-immune-cells-in-solid-tumors/.