Although genomic (DNA) and transcriptomic (messenger RNA) data are easier to collect and analyze, these are only two of the relevant types of “omic” data that are important for understanding disease and the response to treatments. Proteomic, metabolomic, and lipidomic data are also contain information that can be used diagnostically, prognostically, in developing treatment strategies, and in monitoring the response to treatment.
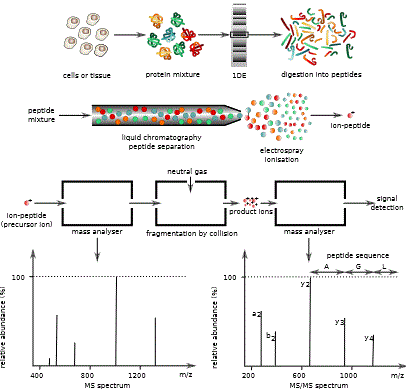
Mass spectrometric approaches enable researchers to look beyond the easily characterized and known molecules.
A criticism of omic mass spectrometry studies is that the experiments are simply “fishing” expeditions that produce large quantities of data, some of which is of dubious quality. Certainly, early studies that used this method had high rates of both false positives (inaccurate assignment of the presence of a molecule that is not really in the sample) and false negatives (failure to reliably detect a molecule present in the sample). Nevertheless, as the methods have evolved and prices have decreased enabling replicate analysis of multiple samples, the ability to generate reliable data through mass spectrometric analysis has increased. Furthermore, researchers have devised strategies to leverage the untargeted nature of mass spectrometry to ask ever more specific questions.
Researchers have devised strategies to leverage the untargeted nature of mass spectrometry to ask ever more specific questions.
The studies highlighted in the commentary articles use mass spectrometry-based proteomic analysis in targeted ways. Only one of them could be construed as a “data dump” and even that example illustrates the power of having a large enough dataset to perform statistical analysis, integrate the proteomic data with other types of biological data, and investigate properties of the proteomic data to make clinically relevant discoveries. As the techniques advance for analyzing the proteome, metabolome, and lipidome, not only will our understanding of basic biology improve but these approaches may become clinically useful for disease diagnosis, predicting the optimal treatment strategy, and then monitoring the response to treatment.
The commentary in the series on the power of proteomics illustrates examples of the applications of proteomic analysis to make clinically important discoveries and provide insights into basic biology. In the commentary related to heart failure, proteomic analysis led researchers to focus on a single target. In the commentary related to vascular biology, proteomics led to the identification of biomarkers of disease progression. In the commentary related to antigen recognition, proteomic analysis will enhance vaccine development. In the commentary about the protein interaction network, the analysis provides mechanistic insights about mutations that cause disease. In the commentary about cancer, the analysis provides information that will help guide the development of new anticancer immunomodulatory treatments and predict those patients most likely to respond to immune checkpoint inhibitor therapy.
N. R. Gough, Proteomic Analysis Leads to Insights into Heart Failure. BioSerendipity (20 June 2017). https://www.bioserendipity.com/2017/06/20/proteomic-analysis-leads-to-insights-into-heart-failure/.
N. R. Gough, The Power of Proteomics in Vascular Disease. BioSerendipity(19 June 2017). https://www.bioserendipity.com/2017/06/19/the-power-of-proteomics-in-vascular-disease/.
N. R. Gough, Proteomics improves antigen prediction. BioSerendipity(14 June 2017) https://www.bioserendipity.com/2017/06/14/proteomics-improves-antigen-prediction/.
N. R. Gough, The Power of Proteomics in Building the Human Protein Interaction Network. BioSerendipity(23 June 2017) https://www.bioserendipity.com/2017/06/23/the-power-of-proteomics-in-building-the-human-protein-interaction-network/.
N. R. Gough, Single-Cell Proteomics Shines Light on the Complexity of Immune Cells in Solid Tumors. BioSerendipity (28 June 2017) https://www.bioserendipity.com/2017/06/28/single-cell-proteomics-shines-light-on-the-complexity-of-immune-cells-in-solid-tumors/.